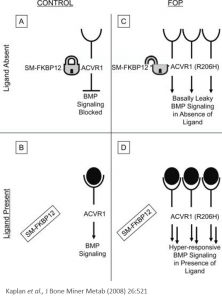
Investigation of ALK2 interactions in Hek293 cells
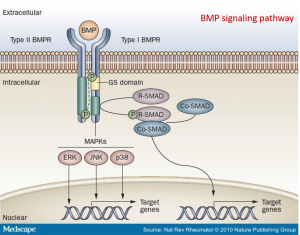
The following BMP4 signal cascade components were chosen for investigation:
- ALK2 wild type receptor
- ALK2 mutated receptors:
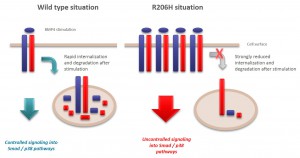
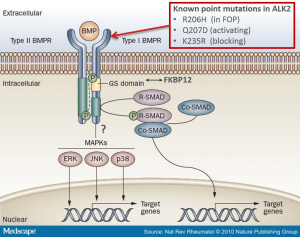
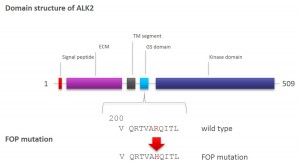
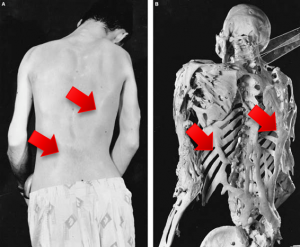
R206H (causes FOP (Fibrodysplasia ossificans progressiva))
Q207D (constitutive active)
K235R (dominant negative)
- ALK6 wild type receptor
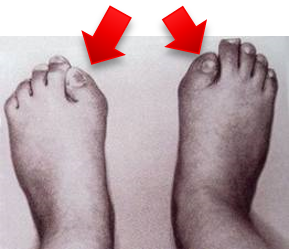
- ALK6 mutated receptor (I200K): causes brachydactyl (shortness of the fingers and toes)
- BMPRII: type II receptor
- FKBP12
Construction of novel expression vectors
Novel expression/purification vectors were generated to stably express the selected baits:
- Strep II and HA epitope tags for detection and purification
- Inducible tet promoter for regulated expression
- Selection marker for stable cell line generation
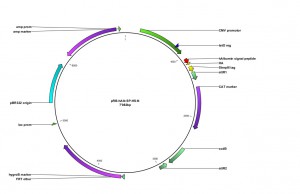
An additional expressionvector was constructedfortheexpressionof type I integral membraneproteinscarrying a cleavable N-terminal signalsequence:
Expression constructs

Transfection and establishment of stable Hek293 cells
- Hek293 cells were transfected with the following constructs using the CaPo method:
ALK2wt (wild type)
ALK2 Q207D
ALK2 K236R
ALK2 R206H
- Stable lines were selected using hygromycine
- Expressed bait proteins were detected by Western blotting using an antibody against the HA tag
- Extraction and purification were optimized using different detergent mixes to ensure optimal recovery of transmembrane receptors
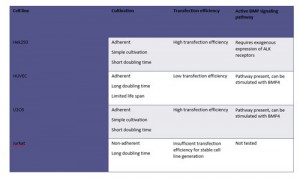
- Establishment of 8 stable Hek293 lines expressing key components of the BMP signaling cascade
- Optimization of purification procedure to ensure reproducible capture of interactors
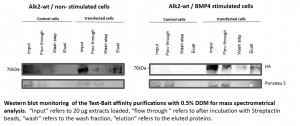
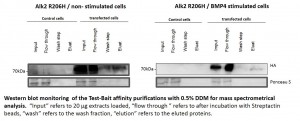
- Western blots of representative purifications are shown below
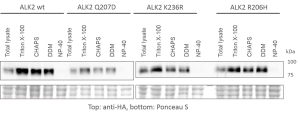
- Optimization of purification procedure to ensure reproducible capture of interactors
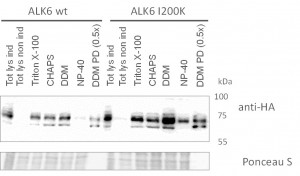
- Western blots comparing purification of ALK6 wild type (wt) and I200K mutant under different conditions from stable Hek293 lines
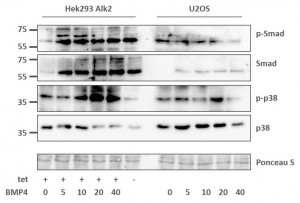
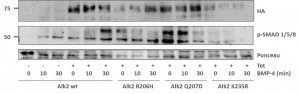
Assessing the BMP signaling pathway in stable Hek293 lines
- Stable Hek293 lines were stimulated with the ligand BMP4 for 30 mins
- Phosphorylation of Smad proteins was assessed using a phospho-specific antibody at the indicated time points
- Stimulation was compared across the ALK2 wild type and mutant receptors
- „Tet“ indicates whether expression of the receptor was induced by tetracycline (+) or whether uninduced cells were used (-)
Purification of receptor complexes from stable Hek293 lines
- Protein complexes around the bait proteins were purified from stable Hek293 lines using the optimized large scale affinity purification procedure
- All purifications were carried out in triplicate using the Strep tag fused to the baits
- Purified protein complexes were digested with trypsin and peptides were subjected to Nano-LC separation, followed by tandem mass spectrometry analysis on a LTQ Orbitrap
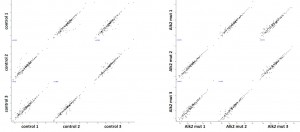
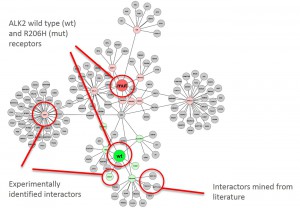
- Triplicate datasets were analyzed using a custom bioinformatics pipeline and interaction networks were generated using the software Cytoscape
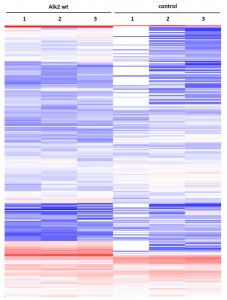
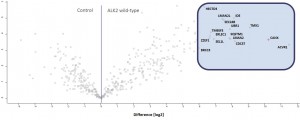
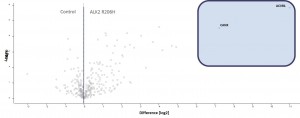
Example of an interactor list generated from a purification of ALK2 wt
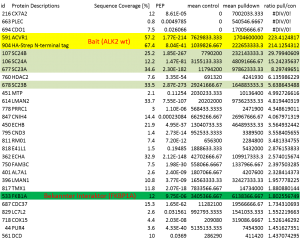
Summary of purifications carried out on 8 stable Hek293 lines
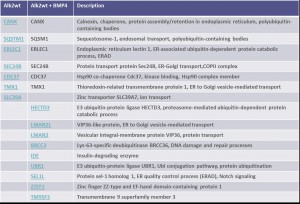
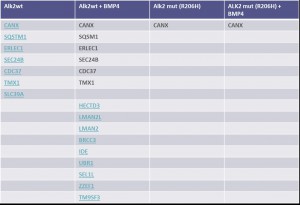
Identifiedinteractorsarelisted in thetablebelow
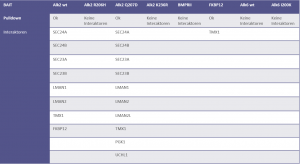
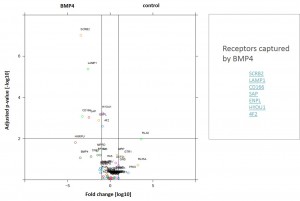
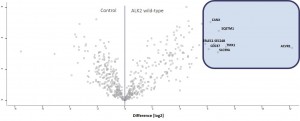
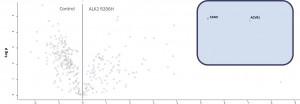
- ALK2 wild type purifications yielded several known and novel interactors
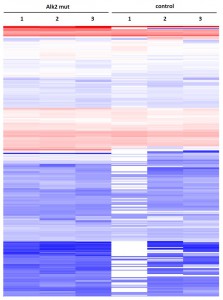
- ALK2 Q207D purifications yielded several interactors which overlap substantially with ALK2 wild type interactors
- No interactions were identified for ALK2 R206H and K236R, or the BMPRII and ALK6 receptors
- FKBP12 purifications yielded one novel interactor, TMX1
- Interactors identified for ALK2 are mostly components involved in receptor sorting, turnover and receptor internalization
Graphical representation of ALK2 interactions in Hek293 cells
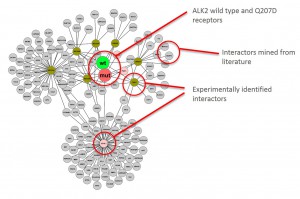
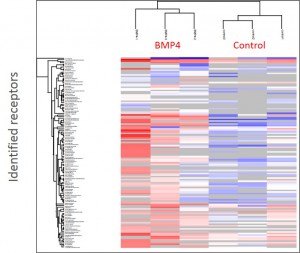
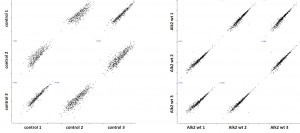
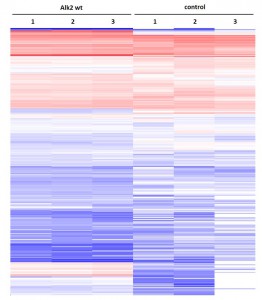
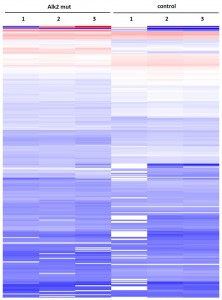
 Cluster analysisof LRC datasets
Cluster analysisof LRC datasets